Duplicate and edit a construct for subcloning
Add, edit or replace a sequence in a vector backbone via the advanced sequence editor to create a final construct ready for synthesis
Use our advanced sequence editing tools to efficiently subclone your gene of interest into your vector backbone.
Contents
1. Duplicate your parent vector
2. Access the sequence editor
3. Subclone you gene of interest
3.1 Replace the multiple cloning site
3.2 Add your custom sequence
4. Further readings
1. Duplicate your parent vector
Duplicating the parent vector and create a copy of your original vector that is linked to the original parent sequence. Doing so means you can keep the original sequence untouched as reference or for future use.
Head to your Constructs collection.
Search for the vector backbone via the search bar.
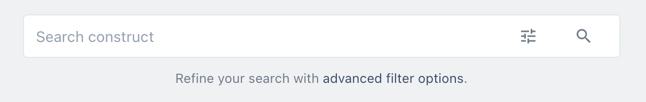
Choose the more options icon ![]() at the top right of the item card.
at the top right of the item card.
Click duplicate to create a copy of you vector.
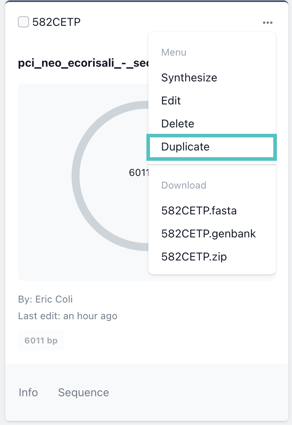
You can update the name and description prior to confirming the duplication.
If you haven't uploaded your vector backbone already, see this article on how to upload a sequence: Upload your vector
2. Access the sequence editor
Now that you have duplicated the parent vector you can edit the copy to insert your gene of interest. Click the construct seq. button in the top right hand of the page to move to the editor.
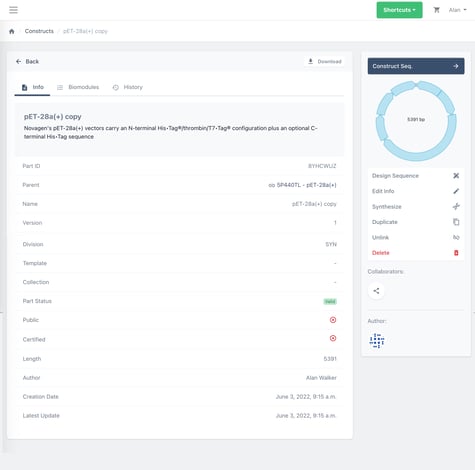
3. Subclone you gene of interest
You have two options for subcloning your gene of interest:
i) you can either replace the multiple cloning site with the sequence of your gene of interest, or
ii) add its sequence to a specific position in the destination vector
3.1 Replace the multiple cloning site
Let's look look at how we can subclone your gene of interest by replacing the multiple cloning site.
First, select the portion of your sequence you'd like to replace. If, for example, you want to subclone your gene of interest between the NdeI and XhoI restriction sites (for more on selecting sequences, see this article)
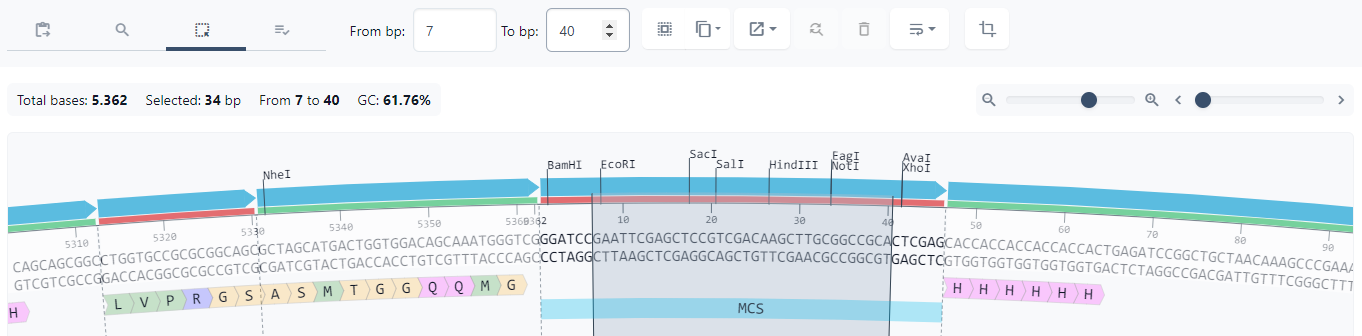
Next, look for the 'replace bases' icon in the toolbar. Then click it.
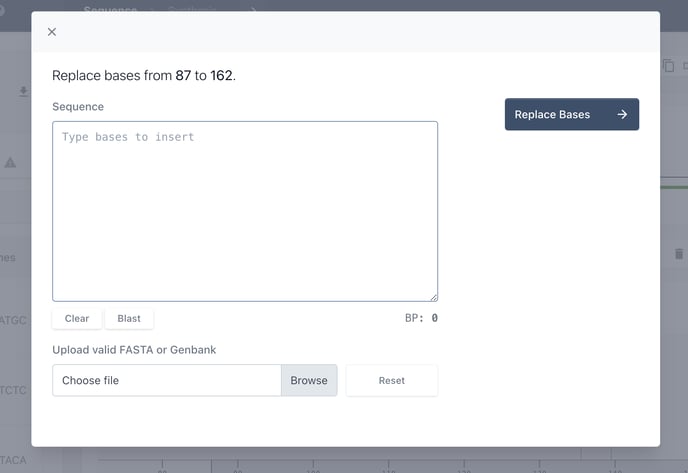
Now you can either copy and paste your desired sequence, or upload a GB file.
3.2 Add your custom sequence
Alternatively, you can add your sequence to the construct without making changes to the original vector. To do this, we will use the add sequences tool:
Navigate to the location of your sequence where you would like to add your transgene.
Click between the two base pairs at the point you would like to add the sequence.
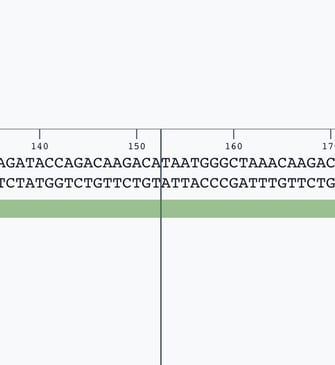
Look for the 'insert bases' tool in the toolbar above the sequence editor.
You can choose between typing you sequence, upload a sequence via a GB file or choose from within you collections.
4. Further readings
More on adding and replaces bases in this article: The Sequence Editing tools
The fastest and simplest way to make these kinds of changes in the Breeze BioCAD is by embracing the modular editing tools. Want to learn more about how to work with modularity? Here you go: Working with Modularity