Create a linear DNA template for in vitro transcription
Breeze BioCAD supports modular design of DNA templates for in vitro transcription of mRNA utilising standard biomodules for CleanCap and precise length Poly A tails.
Contents
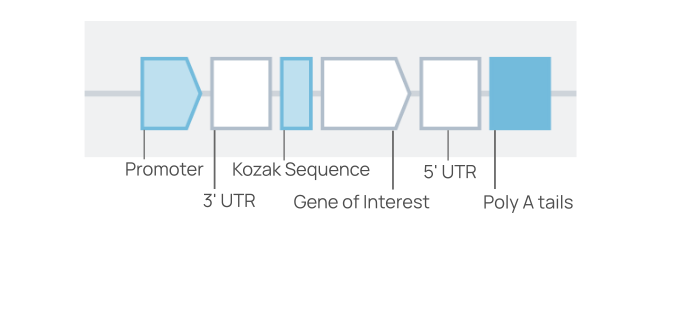
The DNA to RNA template structure
The Linear DNA template for In Vitro Transcription contains six placeholders and three biomodules intended to support construct design for transcription with clean cap. Three of the placeholders are prefilled with suitable biomodules for the transcription protocol. These three biomodules (shown below in blue) can be changed according to your designs.
1) Create a New Construct
Navigate to the Construct page
Choose to create a new construct
Create a new construct from scratch
Give your construct a name and choose a starting template you would like to begin with. We will change this in the next step.
2) Choose linear DNA for RNA IVT template
Lets dive into editing the construct so we can change the template and choose the DNA template for RNA.
From the page you were directed to after creating the new construct, hit the Construct Seq. button.
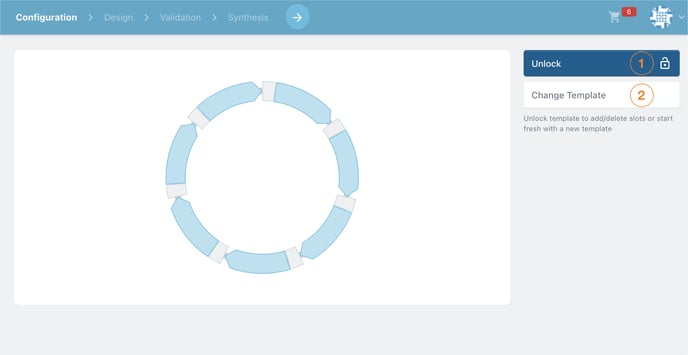
Now look for the pencil icon ![]() in the top left hand side of the screen. Clicking this will take us to the template editor.
in the top left hand side of the screen. Clicking this will take us to the template editor.
![]()
Choose to Change Template (number 2 in the image below).
Choose the RNA template
We can now see we've change the construct template in the subsequent design page.
3) Add and exchange biomodules
Now we've changed the construct template we can begin adding biomodules to the empty placeholders. A good place to start will be with the placeholder for the Gene Of Interest (GOI) biomodule, which will be white.
Clicking this placeholder will turn the placeholder green.
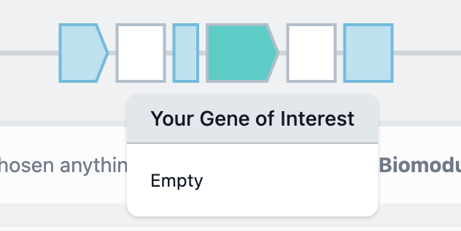
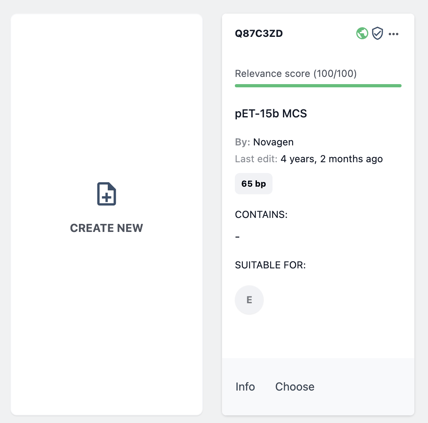
Now you can choose to either add an existing biomodule from your public or private collection, or create a new biomodule by uploading sequence data for your GOI.
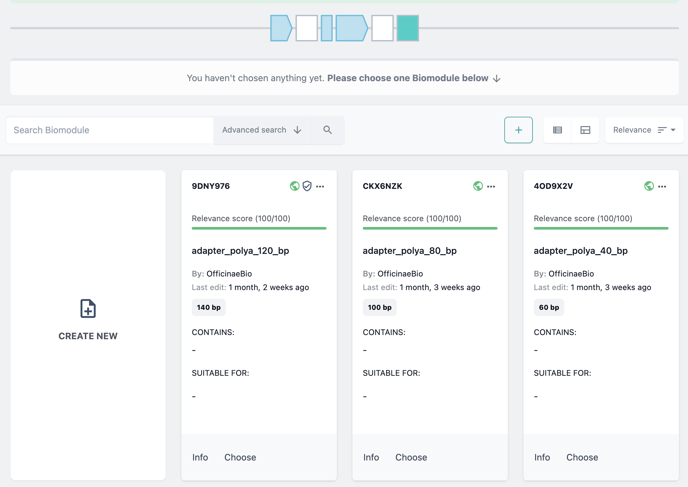
To add a biomodule from your collection click Choose on the biomodule item card.
Alternatively, choose Create New to upload a sequence file for your GOI. Then follow the new biomodule wizard.
For more information on creating biomodules from a file, see this article.
Once you've added a new biomodule, the placeholder will change from green to blue.
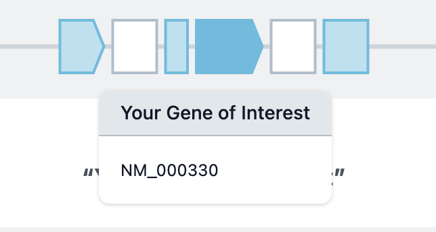
Repeat this process for the 3' and 5' UTR.
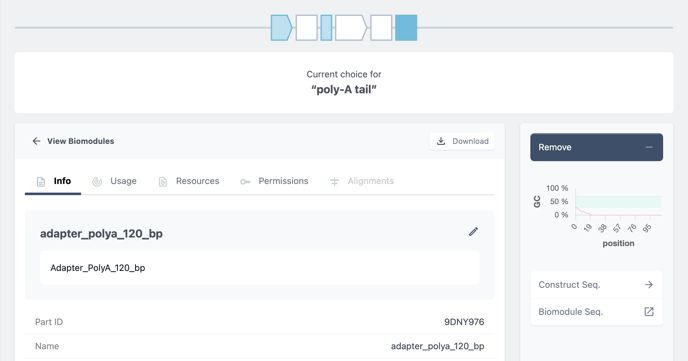
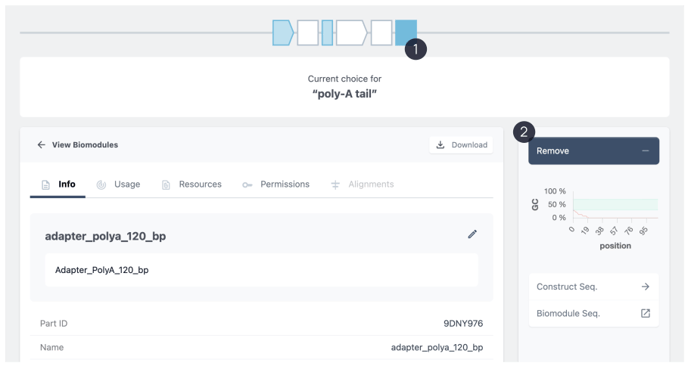
For the prefilled placeholders you can remove and replace the current biomodule with another. A good candidate to exchange the biomodule will be the poly A tail. Here we can exchange exact length Poly A tail biomodules.
To do so, click on the placeholder in blue. (1)
Then choose to remove the biomodule. (2)
You can choose from your biomodule collection or upload your own, just as before.
4) Validate and Synthesise
Now we've create our modular construct for RNA, we can run a checkup for manufacture issues. Just hit Check Up on navigation bar at the top of the screen.

To run a check up on your sequence follow this guide.
Then when you're ready, proceed to synthesis.
Bulk design multiple constructs
You can also design DNA to RNA templates in bulk using the bulk design tool. The process is much the same as that described above.
Learn about bulk designing constructs here: