Annotate a sequence
Quickly create annotations for regulatory regions, UTRs and more with the sequence editor.
Open Sequence Editor
Select sequence
Annotate sequence
Confirm
Step 1: Open the sequence editor
First, you must find the biomodule or construct to which we would like to add the annotation, then view in the sequence editor.
Select sequence on the item card of the biomodule or construct to view it in the sequence editor.
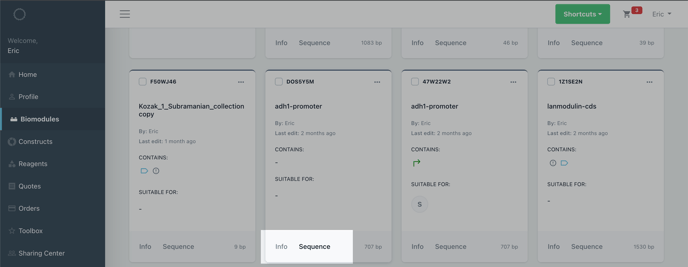
Step 2: Select the sequence you would like to annotate
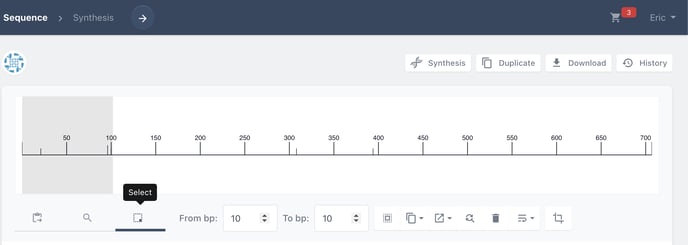
Now we can select the sequence we would like to annotate. Here, we are selecting the entirety of the sequence with the select all tool. Alternatively, you can insert the start and stop positions for your annotation.
Choose select from the toolbar above the sequence viewer.
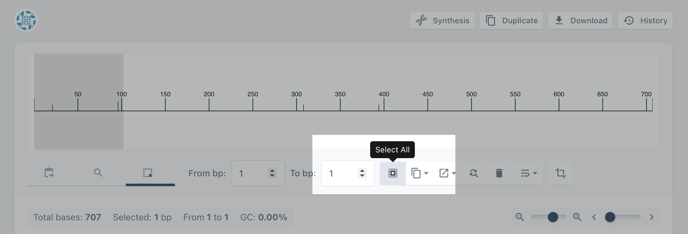
Choose select all to select the entire sequence. Or alternatively, insert the start and stop positions for your annotation
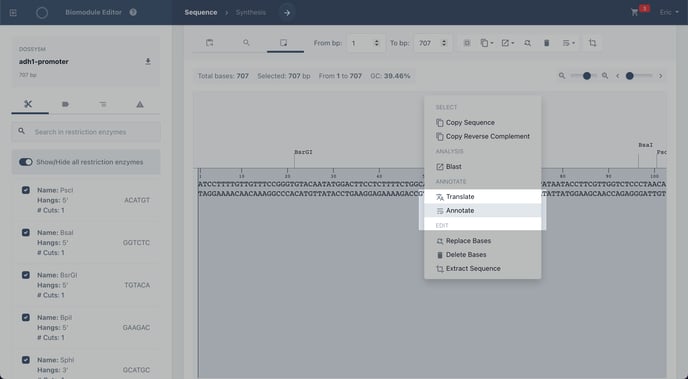
Step 3: Choose the annotate tool
Now we can annotate the sequence we have just selected.
Right click on the sequence to view the editing tools.
Click annotate
Step 4: Complete your annotation
Now we can select the type of annotation and add details to characterise that part of the sequence.
There are a wide range of annotations you can choose from, each with their own features for characterisation. Here, we will annotate a promoter, with features for characterising strength, type, cell affinity, origin and host species.
Choose the type of annotation you would like to add, name it and add a description.
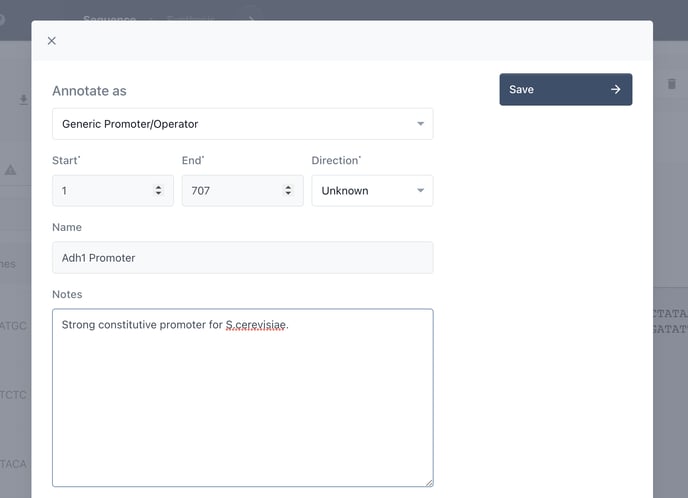
Now add further details where relevant.
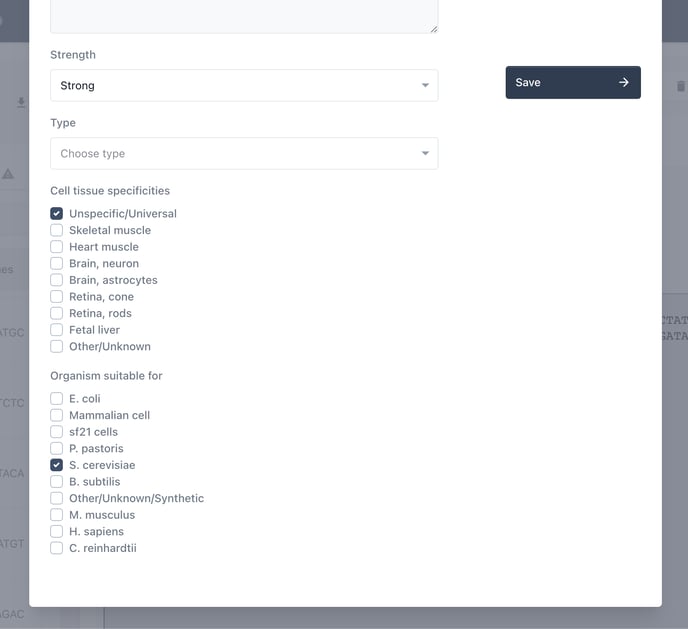
Choose update to save your changes.
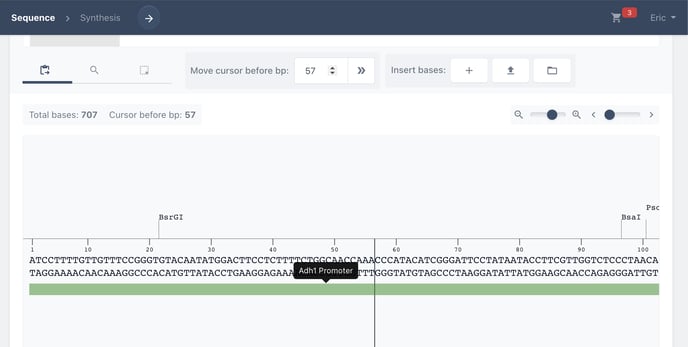
Your annotation is now visible as a coloured bar beneath the sequence.